Task
ML Jähnichen HUB
Compare different ridge regression algorithms. In general, the ridge regression cost function is given by Interpreted as an ordinary optimization problem, we can consider different settings for $\Phi(\cdot)$.
- If Φ(·) is the identity function. we obtain simple linear regression.
- If Φ(·) transforms single $X_{i}$ into a vector of polynomials of $X_{i}$ , we obtain polynomial regression.
- If Φ(·) defines a set of radial basis functions that transform the input $X_{i}$ and the number of bases is equal to the number of examples, we obtain kernel regression.
# satisfying this task we will need
import numpy as np
import matplotlib.pyplot as plt
import time # fun sake
part a
Generate a simple training data set, e.g. take the sine wave and add zero-mean unit-variance Gaussian noise to it, i.e. generate according to $y_{i} = \sin(x_{i}) + \epsilon, \epsilon N(0, 1) $ . Important is, that the underlying function is non-linear.
For fullfilling the task (a) we will create a test and training set with the distribution given in the excercise:
n_train = 5000
n_test = 1000
def data_gen(n, order=False):
"""
Generates test/training set and labels - sinus curve + noise
:param n: size of array
:return: x = np.array(1,n), y = np.array(1,n)
"""
if not order:
x = np.random.uniform(-2 * np.pi, 2 * np.pi, size = n)
else:
x = np.linspace(-2 * np.pi, 2 * np.pi, num = n)
σ = 0.1 * np.random.randn(n)
y = np.sin(x) + σ
return x, y
def exc_a():
# generate training and test set respectively
x_train, y_train = data_gen(n_train, order=False)
x_test, y_test = data_gen(n_test, order=True)
# plot the damn thing
plt.plot(x_train, y_train, 'o', label='train'), plt.plot(x_test, y_test, '.', alpha=0.4, label='test')
plt.legend()
plt.show()
return x_train, y_train, x_test, y_test
s = time.time()
x_train, y_train, x_test, y_test = exc_a()
print("time needed for (a):", time.time()-s, "sec")
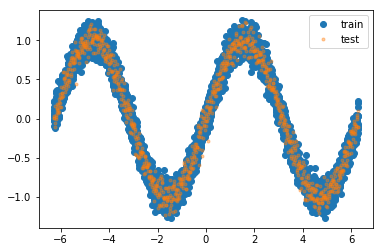
time needed for (a): 0.2388453483581543 sec
preface (a) & (b)
Both the following tasks, ask for exploitation of different regressors. The regressor differ in their designmatrix and hyperparameter for crossvalidation. So we created a linear regressor class, that allows to inherit basic functionalities for linear regression modells with optional adaption of the designmatrix and hyperparameter for subclasses.
class Reg():
def __init__(self, X, y):
"""
Establish a regressor with given data points
:param X: data points, np.array(N, features)
:param y: labels, np.array(N,)
"""
self.θ = np.zeros(1 + 1)
self.δ = 0.005
self.x_train = X
self.X = self.des_mat(X)
self.Y = y.T
def des_mat(self, X):
"""
Converts row vector X=shape(n,) into design matrix
:param X: takes data set as row vector
:return: design matrix np.array((n, 2))
"""
power_cols = np.power(X[:, None], np.arange(1 + 1))
return power_cols
def LSE(self):
"""
Calculates least square solution and saves it in self.θ
"""
A = np.dot(self.X.T, self.X) + self.δ * np.eye(self.θ.shape[0])
b = np.dot(self.X.T, self.Y)
self.θ = np.linalg.solve(A, b)
def predict(self, X):
"""
Predicts values for inserted data row
:param X: data set as row vector np.array(1,n)
:return: row vector np.array(1,n)
"""
X = self.des_mat(X)
pred = np.dot(X, self.θ)
return pred.T
def error(self, X, y):
"""
Measures accuracy of prediction vs. labels via mean squared error
:param X: data set as row vector np.array(1,n)
:param y: labels as row vector np.array(1,n)
:return: float, accuracy level
"""
n = len(X)
pred = self.predict(X).T
Y = y.T
err = 1 / n * np.linalg.norm(Y - pred)
return err
Additionally the following task ask for crossvalidation, with is achieved by the following method. Please note that the regressor is exchangable.
def cross_validation(Δ, X, y, k=5, regressor=PolReg):
"""
k-fold cross validation for polynomial regression or kernel regression
:param Δ: tuple (float) of polynomial degrees or lambdas
:param X: data points, np.array(N, features)
:param y: labels, np.array(N,)
:param k: integer that specifies the foldedness of the cross validation
:param regressor: child of Reg with existing parameter for cross-validation
:return: np.array filled with mean accuracy
"""
# placeholder for evaluating cross validation
eva = np.zeros(shape=(k, len(Δ)))
# we must shift the indices for 5 fold below
shifter = int(1 / k * len(X))
for j in range(k):
for i, h in enumerate(Δ):
shift = j * shifter
# get local training and test set
_x, _y = map(lambda d:
np.roll(d, shift),
[X, y])
_x_train, _y_train = map(lambda d:
d[shifter:],
[_x, _y])
_x_test, _y_test = map(lambda d:
d[:shifter],
[_x, _y])
_reg = regressor(_x_train, _y_train, h)
_reg.LSE()
eva[j, i] = _reg.error(_x_test, _y_test)
return np.mean(eva, axis=0)
Finaly, also the visualization is invariant of its regressor:
def cross_val_viz(Δ, err, x_test, y_test, y_reg):
fig = plt.figure()
ax1 = fig.add_subplot(211)
ax1.set_title("crossvalidation")
plt.plot(Δ, err, label='error')
plt.legend()
ax2 = fig.add_subplot(212)
ax2.set_title("regression")
plt.plot(x_test, y_test, 'o', label='train'), plt.plot(x_test, y_reg, label='regression')
ax2.legend()
plt.show()
part b
Fit a polynomial regression to the data set and visualize its predictions. Determine the degree by cross-validation.
First of all we need to inherit basic functionalities, add hyperparameter and adapt the designmatrix with respect to polynomial regression. This is achieved with the following class:
class PolReg(Reg):
"""
Instantiate a polynomial regressor with given data points
:param X: data points, np.array(N, features)
:param y: labels, np.array(N,)
"""
def __init__(self, X, y, deg=1):
self.deg = deg
Reg.__init__(self, X, y)
self.θ = np.zeros(self.deg + 1)
def des_mat(self, X):
"""
Converts row vector x=shape(1,n) into polynomial regression design matrix
:param X: takes data set as row vector
:return: design matrix np.array((n, deg +1)) or np.array((n,len(µ))
"""
power_cols = np.power(X[:, None], np.arange(self.deg + 1))
return power_cols
def exc_b():
# choose the range of values for crossvalidation
Δ = np.arange(30)
# get accuracies from the different Δ values
err = cross_validation(Δ, x_train, y_train, k=5, regressor=PolReg)
# get the best index
bst_deg = Δ[np.argmin(err)]
print("Best degree for polynomial regression: ", bst_deg)
# solve least square solution and get prediction
_pol = PolReg(x_train, y_train, deg=bst_deg)
_pol.LSE()
y_reg = _pol.predict(x_test)
# plot results
cross_val_viz(Δ, err, x_test, y_test, y_reg)
s = time.time()
exc_b()
print("time needed for (b):", time.time()-s, "sec")
Best degree for polynomial regression: 11
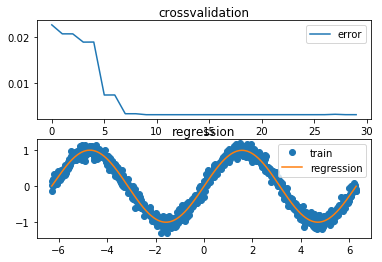
time needed for (b): 2.10097336769104 sec
part c
Fit a kernel regression to the data. Use radial basis functions of the form for each $X_{i}$ consider basis functions $\kappa$ with means $\mu_{j}=X_{j}$ , j = {1, … , n}. Determine λ using cross-validation and visualize your results.
So same, same but different regressor
class KerReg(Reg):
def __init__(self, X, y, λ=1, µ=np.linspace(-2 * np.pi, 2 * np.pi, 20)):
"""
Instantiate a kernel regressor with given data points
:param X: data points, np.array(N, features)
:param y: labels, np.array(N,)
"""
self.λ = λ
self.µ = µ
Reg.__init__(self, X, y)
self.θ = np.zeros(len(self.µ))
def des_mat(self, X):
"""
Converts row vector x=shape(1,n) into design matrix
:param X: takes data set as row vector
:return: design matrix np.array((n, deg +1)) or np.array((n,len(µ))
"""
# radial basis function:
# κ(X, µ, λ) = exp(1/λ|| X - µ ||²)
# exp(
X = np.exp(
# 1/λ
-1 / self.λ
# || x - µ ||²)
* np.abs(
X[:,None] - self.µ) ** 2
)
return X
def exc_c():
# choose the range of values for crossvalidation
Δ = np.linspace(5, 20, 30)
# get accuracies from
err = cross_validation(Δ, x_train, y_train, k=5, regressor=KerReg)
# get the best index
bst_λ = Δ[np.argmin(err)]
print("Best lambda for kernel regression: ", bst_λ)
_ker = KerReg(x_train, y_train, λ=bst_λ)
_ker.LSE()
y_reg = _ker.predict(x_test)
# plot results
cross_val_viz(Δ, err, x_test, y_test, y_reg)
s = time.time()
exc_c()
print("time needed for (c):", time.time()-s, "sec")
Best lambda for kernel regression: 10.1724137931
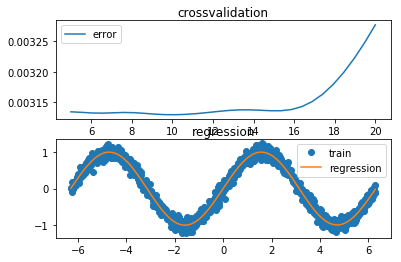
time needed for (c): 1.5013198852539062 sec
Thanks and best
Assion & Greßner
 Music
Music